
主要研究兴趣:
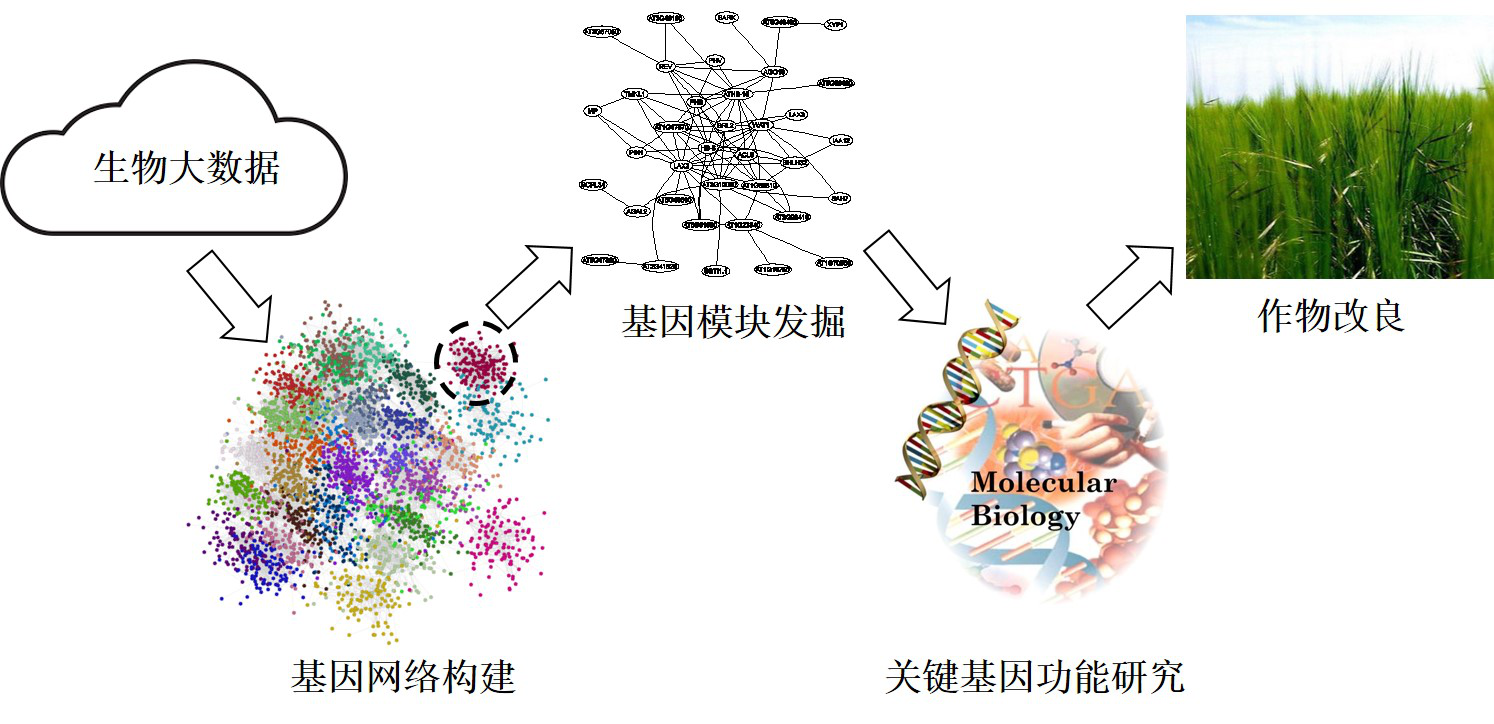
生物大数据的挖掘与应用
基因网络的构建与应用
作物抗逆及养分高效利用基因模块的发掘与功能研究
教授,博士生导师。 2001年毕业于北京大学生命科学学院, 获理学学士学位; 2007年获美国伊利诺伊大学香槟分校生物学博士学位,后在耶鲁大学(2007 – 2010年)和加州大学戴维斯分校(2010 – 2016年)从事研究工作, 任博士后研究员、助理项目科学家等职。
2016年加入中国科学技术大学生命科学学院。多年来一直从事植物系统生物学的研究,致力于分析、整合生物大数据以构建基因表达网络,并从中发掘出作用于植物代谢、发育、逆境应答等各种生命过程的基因模块,为这些过程的后续研究提供了崭新思路;同时也开发了植物蛋白芯片技术,以用于高通量的蛋白功能研究。研究成果发表于 PLoS Genetics, Genome Research, Genome Biology, Plant Communications, Plant Journal, Cell Reports Methods 等期刊上,被 Faculty of 1000 推荐,SCI总引用超过3300次。
代表性论文:
1. Han E, Geng Z, Qin Y, Wang Y, and Ma S. 2024. Single-cell network analysis reveals gene expression programs for Arabidopsis root development and metabolism. Plant Communications 5:100978. [Full text]
2. Xu Y, Wang Y, and Ma S. 2024. SingleCellGGM enables gene expression program identification from single-cell transcriptomes and facilitates universal cell label transfer. Cell Reports Methods 4:100813. [Full text]
3. Peng Y, Liu Y, Wang Y, Geng Z, Qin Y, and Ma S. 2024. Stomatal maturomics: hunting genes regulating guard cell maturation and function formation from single-cell transcriptomes. Journal of Genetics and Genomics 51:1286-1299 [Full text]
4. Wang Y, Zhang Y, Yu N, Li B, Gong J, Mei Y, Bao J, and Ma S. 2023. Decoding transcriptional regulation via a human gene expression predictor. Journal of Genetics and Genomics. [Full text]
5. Zhang Y, Han E, Peng Y, Wang Y, Wang Y, Geng Z, Xu Y, Geng H, Qian Y, and Ma S. 2022. Rice network analysis identified gene modules associated with agronomic traits. Plant Physiology 190:1526-1542 [Full text]
6. Peng Y#, Zuo W#, Zhou H, Miao F, Zhang Y, Qin Y, Liu Y, Long Y, and Ma S. 2022. EXPLICIT-Kinase: a gene expression predictor for dissecting the functions of the Arabidopsis kinome. Journal of Integrative Plant Biology (#共同一作) [Full text]
7. Geng H#, Wang M#, Gong J#, Xu Y, and Ma S. 2021. An Arabidopsis expression predictor enables inference of transcriptional regulators for gene modules. Plant Journal 107:597-612. (#共同一作) (Plant Journal 杂志2021年Outstanding Paper Award)[Full text]
8. Lee D, Lal NK, Lin ZD, Ma S, Liu J, Castro B, Toruno T, Dinesh-Kumar SP, and Coaker G. 2020. Regulation of reactive oxygen species during plant immunity through phosphorylation and ubiquitination of RBOHD. Nature Communications 11:1838.
9. Shen Y, Liu J, Geng H, Zhang J, Liu Y, Zhang H, Xing S, Du J*, Ma S*, and Tian Z*. 2018. De novo assembly of a Chinese soybean genome. Science China Life Sciences 61:871-884. (*co-corresponding author) (*共同通讯作者)
10. Ma S*, Ding Z, Li P*. 2017. Maize network analysis revealed gene modules involved in development, nutrients utilization, metabolism, and stress response. BMC Plant Biology 17:131. (*共同通讯作者)
11. Feng B, Ma S, Chen S, Zhu N, Zhang S, Yu B, Yu Y, Le B, Chen X, Dinesh-Kumar SP, Shan L, He P. 2016. PARylation of the forkhead-associated domain protein DAWDLE regulates plant immunity. EMBO Reports 17:1799-1813.
12. Ma S*, Bohnert HJ, Dinesh-Kumar SP*. 2015. AtGGM2014, an Arabidopsis gene co-expression network for functional studies. Science China Life sciences 58: 276-286. (*共同通讯作者)
13. Ma S*, Shah S, Bohnert HJ, Snyder M, Dinesh-Kumar SP*. 2013. Incorporating Motif Analysis into Gene Co-expression Networks Reveals Novel Modular Expression Pattern and New Signaling Pathways. PLOS Genetics 9: e1003840. (*共同通讯作者)
14. Padmanabhan MS, Ma S, Burch-Smith TM, Czymmek K, Huijser P, Dinesh-Kumar SP. 2013. Novel Positive Regulatory Role for the SPL6 Transcription Factor in the N TIR-NB-LRR Receptor-Mediated Plant Innate Immunity. PLOS Pathogens 9: e1003235.
15. Ma S*, Bachan S, Porto M, Bohnert HJ, Snyder M, Dinesh-Kumar SP*. 2012. Discovery of stress responsive DNA regulatory motifs in Arabidopsis. PLOS One 7: e43198. (*共同通讯作者)
16. Lee HY, Bowen CH, Popescu GV, Kang H-G, Kato N, Ma S, Dinesh-Kumar S, Snyder M, Popescu SC. 2011. Arabidopsis RTNLB1 and RTNLB2 Reticulon-Like Proteins Regulate Intracellular Trafficking and Activity of the FLS2 Immune Receptor. Plant Cell 23: 3374-3391.
17. Ma S, Bohnert HJ. 2007. Integration of Arabidopsis thaliana stress-related transcript profiles, promoter structures, and cell-specific expression. Genome Biology 8: R49.
18. Ma S, Gong Q, Bohnert HJ. 2007. An Arabidopsis gene network based on the graphical Gaussian model. Genome Research 17: 1614-1625.
19. Bohnert H, Gong Q, Li P, Ma S. 2006. Unraveling abiotic stress tolerance mechanisms - getting genomics going. Current Opinion in Plant Biology 9: 180-188.
20. Ma S, Gong Q, Bohnert H. 2006. Dissecting salt stress pathways. Journal of Experimental Botany 57: 1097-1107.
21. Gong Q, Li P, Ma S, Rupassara S, Bohnert H. 2005. Salinity stress adaptation competence in the extremophile Thellungiella halophila in comparison with its relative Arabidopsis thaliana. Plant Journal 44: 826-839.
谷歌学术页面:http://scholar.google.com/citations?user=fpVHlecAAAAJ
招生招聘:实验室常年招聘有植物分子生物学和生物信息学相关背景的博士后和实验员,待遇从优。同时热忱欢迎本科生前来实习或攻读硕士、博士学位。
联系方式:sma@ustc.edu.cn
实验室网址:https://faculty.ustc.edu.cn/mashisong/zh_CN/index.htm
ResearchID: http://www.researcherid.com/rid/E-7225-2012
